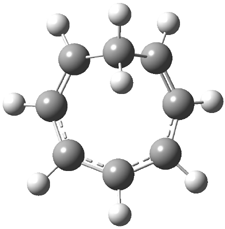
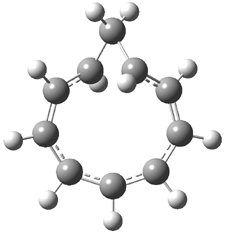
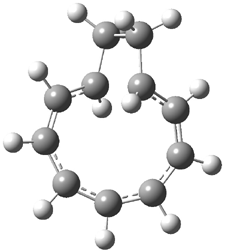
Here’s another interesting application of computed NMR spectra to resolve the structure of natural products. Braddock and Rzepa have examined obtusallenes V (1), VI (2) and VII (3).1 The geometries were optimized at mPW1PW91/6-31G(d,p) and the chemical shifts were obtained at this level and using the aug-cc-pVDZ basis set. The larger basis reduces the error and no statistical correction need be applied. The coordinates of these compounds are available through this web-enhanced object of the paper.
|
|
|
|
The confusion in these structures relates to the position of the halide attachments. For 1 and 2, the problem is which halide (Br or Cl) is at C-7 and C-13. The original structures proposed had these halogens switched from what I’ve drawn, and the correlation between the computed chemical shifts for these original structures and the experiment shows significant deviation: a mean deviation of 1.42 ppm for 1 and 1.67 ppm for 2. Using the structures shown above, along with switching the assigned 13C chemical shifts gives much better agreement between the computed and experimental values; the mean deviation is 1.15 ppm for both 1 and 2. Unfortunately the stereochemistry about the allene cannot be determined using NMR – the two different isomers have similar chemical shifts. Similarly, the structure of 3 is predicted as shown above, though the experiment reported only some of the chemical shifts so some uncertainty remains.
References
(1) Braddock, D. C.; Rzepa, H. S., "Structural Reassignment of Obtusallenes V, VI, and VII by GIAO-Based Density Functional Prediction," J. Nat. Prod., 2008, DOI: 10.1021/np0705918.
InChIs
1: InChI=1/C15H18Br3ClO3/c1-8-14(18)12-6-13(17)15(22-12)7-10(19)11(21-15)5-9(20-8)3-2-4-16/h3-4,8-14H,5-7H2,1H3/t2-,8-,9+,10+,11-,12+,13-,14-,15-/m0/s1
InChIKey = PVIUYMGCQVXTIT-JUHTWQEGBT
2: InChI=1/C15H19Br2ClO3/c1-9-14(17)12-4-5-15(20-12)8-11(18)13(21-15)7-10(19-9)3-2-6-16/h3,6,9-14H,4-5,7-8H2,1H3/t2-,9-,10+,11+,12+,13-,14-,15-/m0/s1
InChIKey = WPEZFVRVOYPLJW-LXJGPXSEBA
3: InChI=1/C15H20Br3ClO3/c1-8-15(18)14-6-10(17)13(22-14)7-11(19)12(20)5-9(21-8)3-2-4-16/h3-4,8-15,20H,5-7H2,1H3/t2-,8-,9+,10-,11+,12-,13-,14+,15-/m0/s1
InChIKey = QTZNVLUNNGQAFG-SOAHCKLOBC

Henry Rzepa responded on 25 Mar 2008 at 7:49 am #
There was earlier discussion on this forum that the Web enhanced objects, being part of the full article, were available only to subscribers. As a result I have requested in the next article, currently in press, that an exact copy of the WEO also be deposited in the supporting information sections, which are freely available to all. I can do this of course, because I did not release to the publisher the full copyright ownership.
On a different aspect. If one submits a crystal structure to a journal nowadays, one has to submit the corresponding CIF file. Paragon Plus (and probably others) will do an automatic validation of the CIF, and if the validation fails, you cannot proceed with the submission! Which is fantastic; we have been waiting years for this! But the day will come when such smart systems will validate a great deal more than just the syntax of a CIF file. Imagine the day when the system will perform a GIAO-prediction of all compounds for which authors assert structures. The outcome, if it conflicts with the actual reported NMR spectrum, might be interesting. OK, this is controversial (GIAO predictions are not yet that reliable), but given the difficulty almost all journals now have of finding referees who actually do a thorough job, surely we must consider whether machines can do at least some of the refereeing?
Would we (as both authors and readers of articles) be happy with such machine refereeing? We might, since we might think we can trust the articles a tad more than we do now!