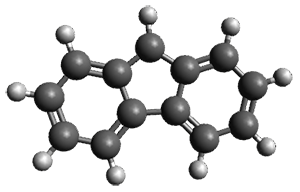
Is the fluorenyl cation 1 antiaromatic or non-aromatic? This is still an open question. But the recent paper by Costa, et al. provides a new path towards potentially answering this question; they have finally synthesized this molecule.1
By photolizing 2 in low-density amorphous ice (LDA ice) and in deuterated ice at 8 K, they have identified a new IR spectrum.
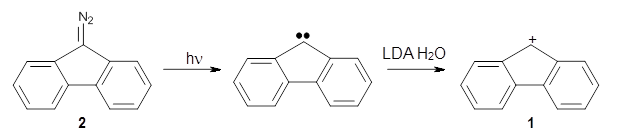
To identify the origin of these spectra, they optimized the geometry of the fluorenyl cation 1 at B3LYP-D3/def2-TZVP (see Figure 1) and computed its IR spectra. These computed IR frequencies were then scaled by 0.97. The agreement between the computed and experimental frequencies is quite reasonable, and the isotopic shifts are also reasonably well reproduced. The agreement is not perfect, as seen in Table 1. Hopefully, further experiments will now be carried out to try to answer the lead question of this post.
Figure 1. B3LYP-D3/def2-TZVP optimized geometry of 1.
Table 1. Experimental and computed IR frequencies (cm-1)
and isotopic shift (in parentheses) of 1.
|
Calc. |
Exp. |
|
1008.8 |
986 |
|
1106.8 (+2.1) |
1076.8 (+1.7) |
|
1152.8 |
1117.2 |
|
1198.6 |
1163.5 |
|
1267.0 |
1235.1 |
|
1373.4 (-16.4) |
1343.7 |
|
1510.8(-8.3) |
1469.0 (-7.3) |
|
1530.7 (-3.2) |
1490.5 (-1.0) |
|
1616.8(-6.4) |
1575.7(-4.4) |
|
1640.9 (0.0) |
1601.2 (-4.0) |
References
(1) Costa, P.; Trosien, I.; Fernandez-Oliva, M.; Sanchez-Garcia, E.; Sander, W. "The Fluorenyl Cation," Angew. Chem. Int. Ed. 2015, 54, 2656-2660, DOI: 10.1002/anie.201411234.
InChIs
1: InChI=1S/C13H9/c1-3-7-12-10(5-1)9-11-6-2-4-8-13(11)12/h1-9H/q+1
InChIKey=KZCNYQVQQBONEY-UHFFFAOYSA-N