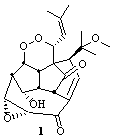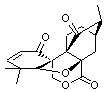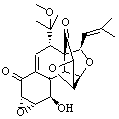In Chapter 1.6.2 we discuss computed NMR spectra, and in particular note some successes in correlating predicted chemical shifts with experiment values. Recently, Rychnovsky took the next logical step, utilizing computational methods to predict the NMR spectrum of a compound whose structure was in doubt.
Hexacyclinol was isolated from Panus Rudis, a type of mushroom. Based on spectroscopic studies, Gräfe proposed 1 as its structure.1 Le Clair claimed to have synthesized a substance with this structure in 2006.2 This article became a cause célèbre in the blogosphere,3 with serious doubts cast upon the veracity of the author and his claims.
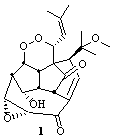
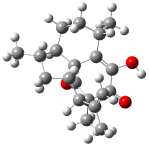
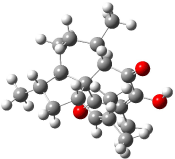
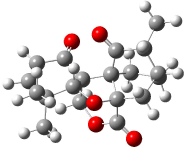
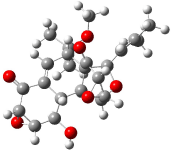
Rychnovsky4 doubted that the molecule actually possessed the unusual structure of 1. Since the actual structure was unknown, he proposed to compute the NMR shifts based on the optimized structure of 1 and compare them with the experimental values. Given the very large size of hexacyclinol, the computational approach would have to be rather limited. Therefore, whatever (small) method was to be employed would have to be tested for adequate predictive performance with known compounds. Rychnovsky selected the three diterpenes elisapterosin B 2, elisabethin A 3, and maoecrystal V 4 to benchmark his computations. His computational approach was to first utilize a Monte Carlo search with the MMFF force field to identify low lying conformers. The best conformer was then optimized at HF/3-21G and the chemical shifts were computed using this geometry with the GIAO/mPW1PW91/6-31G(d,p). The optimized structures of the diterpenes 2-3 are shown in Figure 1.
Figure 1. HF/3-21G optimized structures of 2-3.4
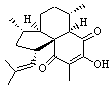
The computed 13C chemical shifts for these test compounds were then plotted against the experimental values and a linear fit was determined to correct the computed values. The average 13C chemical shift difference between computation and experiment is less than 2 ppm, and no difference exceeds 5 ppm. Next, Rychnovsky optimized the proposed structure of hexacyclinol 1, shown in Figure 2, and computed its 13C chemical shifts and corrected them using the fitting procedure developed for the three test compounds. These computed chemical shifts were in poor agreement with the experimental values; the average deviation was 6.8 ppm and five shifts differ by more than 10 ppm. Rychnovsky concluded that this poor agreement discredits the proposed structure 1.
1
xyz file
Figure 2. HF/3-21G optimized structures of 1.4
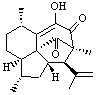
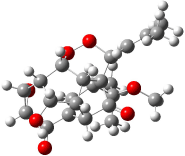
As an alternative, Rychnovsky proposed that hexacyclinol is in fact the by-product from work-up of the natural product panepophenanthrin, also obtained from Panus rudis. He proposed that hexacylinol has the structure shown in 5. He optimized the geometry of 5 and obtained two low-energy conformers. The second-lowest conformer, shown in Figure 3, has a predicted 13C NMR spectrum in very close agreement with experiment. Its average chemical shift deviation is 1.8 ppm with a maximum difference of 5.8 ppm. These differences are consistent with those found in the diterpenes test set. This structure has now been synthesized by Porco and its x-ray structure obtained.5 This compound has the structure predicted by Rychnovsky and is completely consistent with the original hexacyclinol compound reported by Gräfe. This successful resolution of the structure of hexacycliinol should spur further use of computational methods to predict NMR spectra and evaluate chemical structures. ACD has recently applied its method for predicting NMR spectra to the problem of hexacylinol.6 You can read about this on the ChemSpider blog.
Figure 3. HF/3-21G optimized structures of 5.4
References
(1) Schlegel, B.; Hartl, A.; Dahse, H.-M.; Gollmick, F. A.; Gräfe, U.; Dorfelt, H.; Kappes, B., “Hexacyclinol, a New Antiproliferative Metabolite of Panus Rudis HKI 0254,” J. Antibiot. 2002, 55, 814-817.
(2) La Clair, J. J., “Total Syntheses of Hexacyclinol, 5-epi-Hexacyclinol, and Desoxohexacyclinol Unveil an Antimalarial Prodrug Motif,” Angew. Chem. Int. Ed. 2006, 45, 2769-2773, DOI: 10.1002/anie.200504033
(3) (a) Halford, B., “Hexacyclinol Debate Heats Up,” Chem. Eng. News 2006, 84 (31, July 28), 11, http://pubs.acs.org/cen/news/84/i31/8431notw1.html. (b) Love, D. “Hexacyclinol? Or Not?” http://pipeline.corantte.com/archives/2006/06/05/hexacyclinol_or_not.php (c) “Structure Revision of Hexacyclinol”, http://totallynthetic.com/blog/?p=110 (d) Halford, B., “Hexacyclinol Showdown: The Biggest Non-Event at the ACS Meeting”, http://cenonline.blogs.com/sanfrancisco_2006/2006/09/hexacyclinol_sh.html (e) “Hexacyclinol Rides Again”, http://www.healthvoices.com/feed/items/blog_perspective/consultants/pharma/2006/07/3/hexacyclinol_rides_again
(4) Rychnovsky, S. D., “Predicting NMR Spectra by Computational Methods: Structure Revision of Hexacyclinol,” Org. Lett. 2006, 8, 2895-2898, DOI: 10.1021/ol0611346
(5) Porco, J. A. J.; Shun Su, S.; Lei, X.; Bardhan, S.; Rychnovsky, S. D., “Total Synthesis and Structure Assignment of (+)-Hexacyclinol,” Angew. Chem. Int. Ed. 2006, 45, 5790-5792, DOI: 10.1002/anie.200602854
(6) Elyashberg, M. E.; Williams, A. J.; Martin, G. E., “Computer-Assisted Structure Verification and Elucidation Tools in NMR-Based Structure Elucidation,” Prog. Nuc. Mag. Res. Spectrosc., 2007, in press, DOI: 10.1016/j.pnmrs.2007.04.003.
InChI
1: InChI=1/C23H28O7/c1-8(2)6-11-23-10(22(3,4)27-5)7-9-12(21(23)26)13-15(23)18(30-29-11)14(17(13)25)19-20(28-19)16(9)24/h6-7,10-15,17-20,25H,1-5H3/t10-,11+,12?,13?,14?,15?,17-,18?,19+,20+,23?/m1/s1
2: InChI=1/C20H26O3/c1-9(2)14-13-8-11(4)12-7-6-10(3)15-16(21)17(22)19(14,5)18(23)20(12,13)15/h10-14,21H,1,6-8H2,2-5H3/t10-,11-,12+,13-,14-,19+,20-/m0/s1
3: InChI=1/C20H28O3/c1-10(2)8-14-9-12(4)15-7-6-11(3)16-18(22)17(21)13(5)19(23)20(14,15)16/h8,11-12,14-16,21H,6-7,9H2,1-5H3/t11-,12-,14?,15+,16-,20-/m1/s1
4: InChI=1/C19H22O5/c1-10-11-4-7-18(13(10)21)17-9-23-15(22)19(18,8-11)24-14(17)16(2,3)6-5-12(17)20/h5-6,10-11,14H,4,7-9H2,1-3H3/t10-,11-,14-,17?,18-,19+/m1/s1
5: InChI=1/C23H28O7/c1-8(2)6-11-23-10(22(3,4)27-5)7-9-12(15(25)18-17(29-18)14(9)24)13(23)16(28-11)19-20(30-19)21(23)26/h6-7,10-13,15-20,25H,1-5H3/t10-,11+,12?,13?,15+,16+,17-,18-,19-,20?,23-/m0/s1