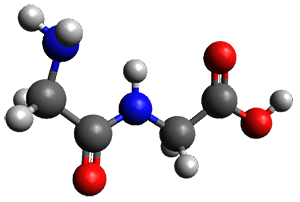
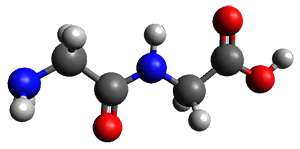
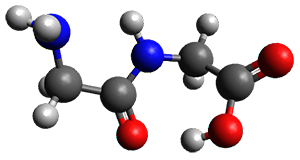
Continuing their application of laser ablation molecular beam Fourier transform microwave (LA-MB-FTMW) spectroscopy and computational chemistry to biochemical molecules (see these previous posts), the Alonso group reports on the structure of the glycine-glycine dipeptide 1.1 The microwave spectrum shows three different conformers. MP2/6-311++G(d,p) computations, the same method they have previously utilized for predicting geometries, revealed a number of different conformations. By matching the spectroscopic parameters obtained from the spectrum with those of the computed structures, they proposed the three conformations 1a, 1b, and 1c, shown in Figure 1.
|
1a |
|
1b |
|
1c |
Figure 1. ωb97xd/6-31G(d) optimized structures of the three conformers of 1.
Note that the authors did not report their structures in their supporting materials(!) so I have optimized them.
The structures of conformers 1a and 1b are nearly planar. MP2 predicts a non-planar rotomer of 1a, which brings the carboxyl group out of plane, to be the lowest conformation in terms of electronic energy. With the M06-2x functional, this non-planar rotomer is about isoenergetic with 1a. With all computational levels 1a is the lowest in free energy. The barrier for rotation between the non-planar rotomer and 1a is very small, and this explains why it is not observed in the supersonic expansion.
References
1) Cabezas, C.; Varela, M.; Alonso, J. L., "The Structure of the Elusive Simplest Dipeptide Gly-Gly." Angew. Chem. Int. Ed. 2017, 56, 6420-6425, DOI: 10.1002/anie.201702425.
InChIs
1: InChI=1S/C4H8N2O3/c5-1-3(7)6-2-4(8)9/h1-2,5H2,(H,6,7)(H,8,9)
InChIKey=YMAWOPBAYDPSLA-UHFFFAOYSA-N

Henry Rzepa responded on 25 Jul 2017 at 1:37 am #
Given the famous postulate by Crick and others in the 1960s that protein structure follows purely from the amino acid sequence, it is of interest to know what the structures of the homopolymers for the 20 principle amino acids are. Thus GlyGly. So it was interesting to find that a crystal structure for GlyGlyGly is known (DOI: 10.5517/cc1k2dwx). Of course the ionisation means it cannot be directly compared to the gas phase structure. The next amino acid alanine is also known as AlaAlaAla (DOI: 10.5517/ccpvzt9). However PhePhePhe appears unknown, as does TyrTyrTyr (although TyrTyrPhe is known; DOI: 10.5517/ccnv43q).
I have not yet tracked down whether any other homotrimers for the 20 amino acids are available as crystal structures. It might also be find to what the longest homopolymer of any amino acid is known as a crystal structure and whether that starts to approach a length that would start exhibiting the tertiary foldings so typical of proteins (I would add that both GlyGlyGly and AlaAlaAla exhibit interesting parallel stackings).
Cn anyone add to the above?