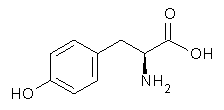
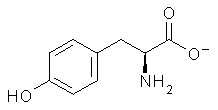
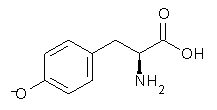
Following on their prediction that the thiol of cysteine1 is more acidic than the carboxylic acid group (see this post), Kass has examined the acidity of tyrosine 1.2 Which is more acidic: the hydroxyl (leading to the phenoxide 2) or the carboxyl (leading to the carboxylate 3) proton?
|
|
|
|
|
|
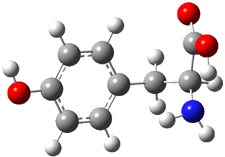
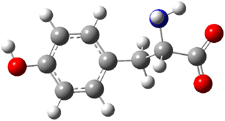
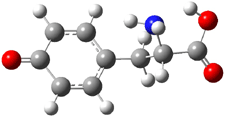
Kass optimized the structures of tyrosine and its two possible conjugate bases at B3LYP/aug-cc-pVDZ, shown in Figure 1, and also computed their energies at G3B3. 2 is predicted to be 0.2 kcal mol-1 lower in energy than 3 at B3LYP and slightly more stable at G3B3 (0.5 kcal mol-1). However, both computational methods underestimate the acidity of acetic acid more than that of phenol. When the deprotonation energies are corrected for this error, the phenolic proton is predicted to be 0.4 kcal mol-1 more acidic than the carboxylate proton at B3LYP and 0.9 kcal mol-1 more acidic at G3B3.
|
1 |
|
|
2 |
3 |
Figure 1. B3LYP/aug-cc-pVDZ optimized structures of tyrosine 1 and its two conjugate bases 2 and 3.2
Gas phase experiments indicate that deprotonation of tyrosine leads to a 70:30 mixture of the phenoxide to carboxylate anions. The computations are in nice agreement with this experiment. (A Boltzmann weighting of the computed lowest energy conformers makes only a small difference to the distribution relative to using simply the single lowest energy conformer.) This demonstrates once again the important role of solvent, since only the carboxylate anion is seen in aqueous solution.
References
(1) Tian, Z.; Pawlow, A.; Poutsma, J. C.; Kass, S. R., "Are Carboxyl Groups the Most Acidic Sites in Amino Acids? Gas-Phase Acidity, H/D Exchange Experiments, and Computations on Cysteine and Its Conjugate Base," J. Am. Chem. Soc., 2007, 129, 5403-5407, DOI: 10.1021/ja0666194.
(2) Tian, Z.; Wang, X.-B.; Wang, L.-S.; Kass, S. R., "Are Carboxyl Groups the Most Acidic Sites in Amino Acids? Gas-Phase Acidities, Photoelectron Spectra, and Computations on Tyrosine, p-Hydroxybenzoic Acid, and Their Conjugate Bases," J. Am. Chem. Soc., 2009, 131, 1174-1181, DOI: 10.1021/ja807982k.
InChIs
1: InChI=1/C9H11NO3/c10-8(9(12)13)5-6-1-3-7(11)4-2-6/h1-4,8,11H,5,10H2,(H,12,13)/t8-/m0/s1/f/h12H
InChIKey=OUYCCCASQSFEME-QAXLLPJCDY
2: InChI=1/C9H11NO3/c10-8(9(12)13)5-6-1-3-7(11)4-2-6/h1-4,8,11H,5,10H2,(H,12,13)/p-1/t8-/m0/s1/fC9H10NO3/q-1
InChIKey=OUYCCCASQSFEME-HVHKCMLZDU
3: InChI=1/C9H11NO3/c10-8(9(12)13)5-6-1-3-7(11)4-2-6/h1-4,8,11H,5,10H2,(H,12,13)/p-1/t8-/m0/s1/fC9H10NO3/h11h,12H/q-1
InChIKey=OUYCCCASQSFEME-XGYCJDCADS

Alex responded on 04 Mar 2009 at 9:25 pm #
cc-pVDZ/B3LYP has been shown to not be accurate to better than 2kcal/mol, so wny should one trust any of this data?
sbachrach responded on 05 Mar 2009 at 9:29 am #
I have three answers to this:
1) The hydroxyl proton is predicted to be more acidic by G3MP2, a method with a smaller error bar.
2) Experiments support the notion of preferential deprotonation of the hydroxyl proton.
3) Just the fact that the two protons are even close in acidity is interesting!